Designs CRISPR protospacers to target single nucleotide polymorphisms with base editors (ABE / CBE). It is free to use. Input required is an rsID or a txt file with rsIDs on each line. https://baglaenkolab.shinyapps.io/snpGuide/ | https://github.com/ybaglaenko/snpGuide
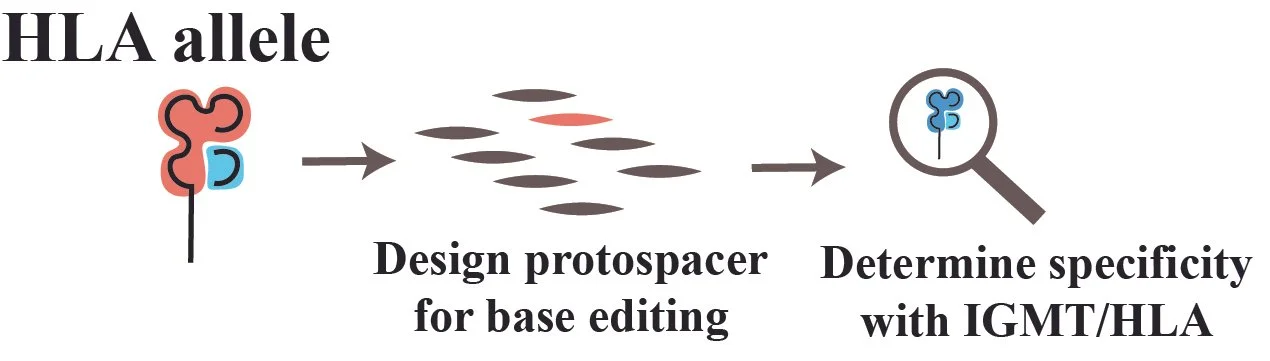
Designs CRISPR protospacers to target or preserve HLA alleles. Example: Ablate HLA-B*27:05 while preserving HLA-B*07:02. It includes plotting, efficiency calculations based on deep learning, and off-target predictions.
A functioning alpha version of HLAguide is available online and hosted by CCHMC. Contact us for use and we will happily share the password while we continue to get the manuscript ready.
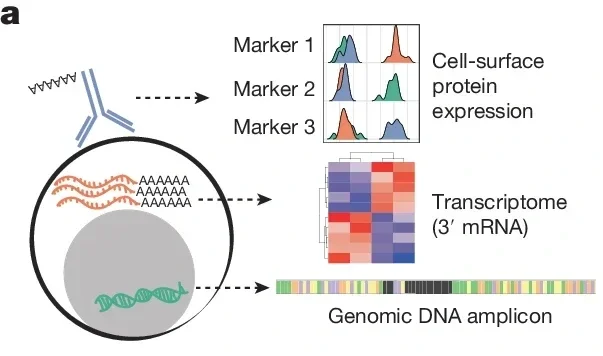
As part of a larger manuscript - we have deposited all analysis codebooks and the full experimental protocol for this work. This is a single cell analysis of genomic DNA amplicons, 3’ RNA sequencing, and ADT sequencing. https://github.com/immunogenomics/craft-seq https://www.protocols.io/view/craftseq-dm6gpz8ojlzp/v2